Introduction:
The development of Bruton tyrosine kinase inhibitors (BTKIs) and their introduction into clinical practice represents a major advance in the treatment of chronic lymphocytic leukemia (CLL). Monotherapy with ibrutinib or other BTKIs generally do not induce complete remissions or undetectable minimal residual disease (uMRD) even with extended therapy. The reason why some patients relapse, and some have minimal residual disease is unknown. Therefore, there is a need to understand the differences between ibrutinib sensitive and resistant CLL cells along with the immune microenvironment to identify novel therapeutic targets.
Methods:
Here, we investigate the cellular heterogeneity of peripheral blood mononuclear cells from patients with CLL treated with ibrutinib using single-cell RNA sequencing. To understand ibrutinib resistance mechanisms, samples were collected from 8 patients at baseline (before treatment) (1), where BTK C481S is present with low variant allele frequency (VAF) (2) and at time of relapse (3). Data from these were compared to ibrutinib sensitive patient samples at baseline (1) (before treatment), 3yr (2) and 5yr (3) on treatment timepoints. Single cells were isolated using the 10X Genomics 5' immune profiling kit. Data were processed with CellRanger v3.1.0. Poor quality cells were filtered out based on a high proportion of mitochondrial reads and low overall feature count. Sample variation was reduced using Batchelor to aid clustering. Clustering was performed using the partitioning and Leiden methods. Cluster top markers were calculated using functions from monocle3. Identity mapping was performed using Seurat and a published PBMC reference dataset.
Results:
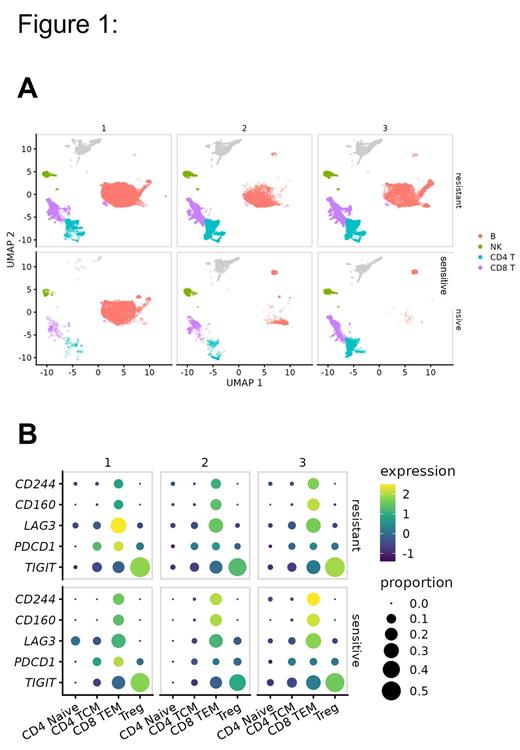
Single-cell RNA sequencing revealed transcriptional heterogeneity within the B cell cluster (Fig 1A). Three subpopulations of B cells were defined based on differential representation in the ibrutinib-sensitive and resistant patients. Ibrutinib sensitive cells showed enrichment of B cell populations with upregulation of MHC I molecules and TNF family members. We also identified that inflammatory response and metabolic-related pathways were decreased, whereas cellular response to stress and DNA repair programs were increased in the ibrutinib resistance samples. Gene module analysis showed that ibrutinib sensitive samples had an increase in T cell activation, RNA splicing, cell adhesion and migration programs during therapy.
We hypothesized that the emergence of BTK C481S mutation in ibrutinib-resistant cells would lead to a distinct transcriptional profile. Long-read sequencing was performed using Oxford Nanopore technology. BTK-mutant cells were identified based on the presence of two or more unique RNA molecules detected with the C481S mutation and these were predominantly identified in the low-VAF and relapse samples. However, there was no significant difference between BTK-mutant cells and the remaining B cells.
To see if T cell phenotypes reflected the differences observed in B cell transcriptional state, we characterized and quantified T cell populations according to ibrutinib response and investigated the changes in the T cell population. Ibrutinib markedly increased CD4+ and CD8+ T cell numbers in both resistant and sensitive CLL patients. However, there was a significant (p<0.05) enrichment of Tregs in the ibrutinib-resistant samples. We found a connection between the immunosuppressive phenotype in the microenvironment demonstrating tumor's intrinsic immunological reactive program with exhausted T cells. Also, we found significant difference in the expression of checkpoint molecules and suppressive genes in T cells (Fig 1B) and NK cells.
Conclusion:
Our findings demonstrate that the B cell population in ibrutinib-resistant and ibrutinib sensitive CLL patients exhibit distinct transcriptional states. At the single-cell level, our findings demonstrate a systematic picture of diverse malignancy and microenvironmental changes in CLL. Overall, these findings provide an insight into the complex molecular and cellular interactions of CLL that may have a crucial role in BTKI resistance.
Disclosures
Rogers:Pharmacyclics: Consultancy; Novartis: Research Funding; AstraZeneca: Consultancy; Beigene: Consultancy; Loxo@Lilly: Consultancy; Janssen: Consultancy; AbbVie: Consultancy, Research Funding; Genentech: Consultancy, Research Funding. Bhat:Aptitude Health: Honoraria; Abbvie: Consultancy; AstraZeneca: Consultancy, Research Funding. Kittai:Abbive: Consultancy; AstraZeneca: Consultancy, Research Funding; BeiGene: Consultancy, Research Funding, Speakers Bureau; Eli Lilly: Consultancy; Janssen: Consultancy; KITE: Consultancy; BMS: Consultancy. Byrd:Newave: Membership on an entity's Board of Directors or advisory committees, Research Funding; Kurome: Current equity holder in publicly-traded company, Membership on an entity's Board of Directors or advisory committees; Vincerx: Current equity holder in publicly-traded company, Membership on an entity's Board of Directors or advisory committees; OSU Drug Devel. Inst.: Consultancy; Orbimed: Consultancy, Research Funding; Eilean Therapeutics: Consultancy, Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees, Research Funding; Orange Grove Bio: Membership on an entity's Board of Directors or advisory committees; American Cancer: Membership on an entity's Board of Directors or advisory committees; AstraZeneca: Other: TRAVEL, ACCOMMODATIONS, EXPENSES. Blachly:AstraZeneca: Consultancy; Epigenetic classification of leukemia: Patents & Royalties: PCT conversion filed; AbbVie: Consultancy; Leukemia Diagnostic Device: Patents & Royalties: Being prosecuted; Astellas: Consultancy. Woyach:Newave: Consultancy; Loxo: Consultancy; Beigene: Consultancy; AstraZeneca: Consultancy; Abbvie: Consultancy; Schrodinger: Research Funding; Morphosys: Research Funding; Karyopharm: Research Funding; Janssen: Consultancy, Research Funding; Pharmacyclics: Consultancy, Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal